Deconvoluting the pooled samples
The pooled samples feature table
Users are free to use their preferred preprocessing software. As mentioned before, it is required that users adopt row/column naming conventions seen in the preparation grids (e.g., grid_1_column_1). A flat feature table (.csv) is necessary to provide to the MultiplexMS app. It is recommended that MS feature tables be in a specific format where features are designated 1 (present - above the minimum intensity threshold) or 0 (absent - zero, or below the minimum intensity threshold). Users can upload a MS feature table containing intensity values for each sample, however, the file create following computational deconvolution will only contain 1 (present) and 0 (absent). In both cases, choosing a sensible intensity cut-off value before processing the table is strongly advised.
The table should show which MS features are present in each pooled sample. The pooled samples feature table might be arranged so that all samples appear as columns and all features in rows. However, this is up to the user.
Feature names do not need to follow any specific pattern. A typical feature table would look like this:
The Deconvolution tab in the companion tool
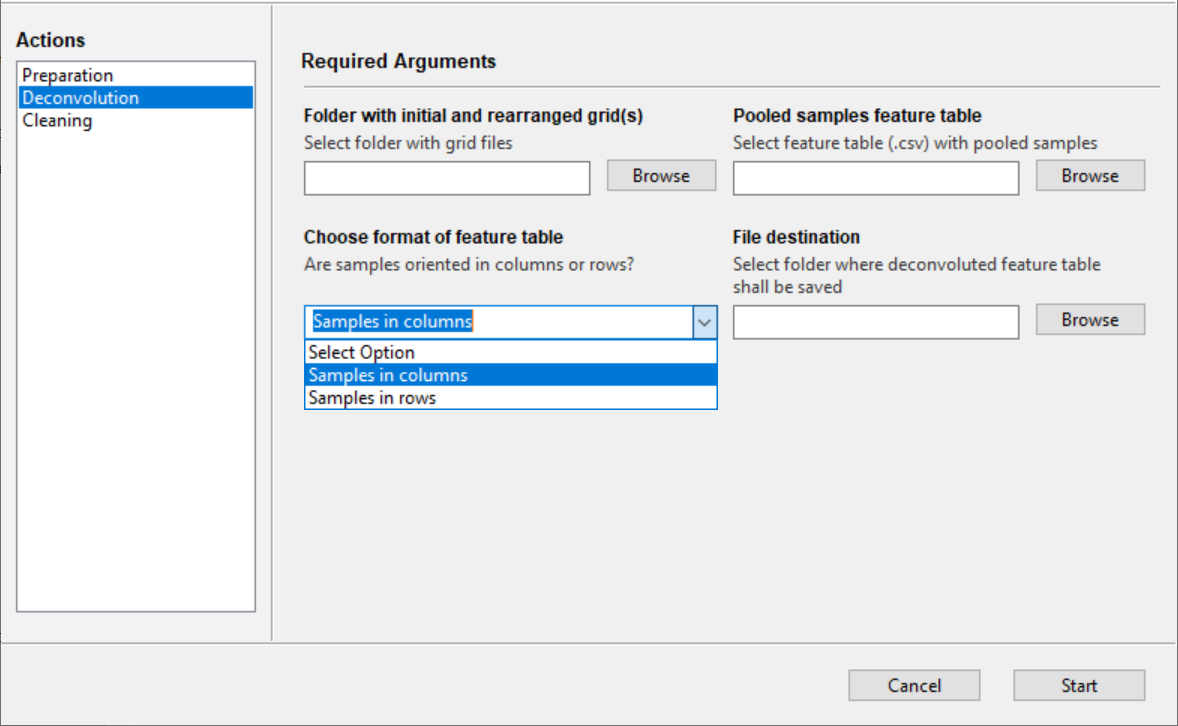
Folder with initial and rearranged grid(s) - Direct the companion tool to the grids folder that was generated by the Preparation function.
Pooled samples feature table - Load the CSV feature table from the MS analysis of the pooled samples. It is important the feature table be in the same format as shown above where pooled samples appear in columns and MS feature are in rows. The opposite arrangement of pooled sample names and features is permitted, where pooled sample names appear in rows and features are in columns - the choice of format is specified by the user in Choose format of feature table.
NOTE: Sample names need to match the pooled sample names in the grids folder. Any prefixes or suffixes need to be deleted prior to submission (e.g., 20220506_grid_1_column_1_R1 should be grid_1_column_1).
Choose format of feature table - The format of the feature table has to be determined. Specify if pooled sample names are oriented in columns (-> features in rows) or if pooled sample names are in rows (-> features in columns).
File destination - Folder where the deconvoluted feature table shall be saved. The deconvoluted feature table will show which MS features are present in the individual samples. Only feature presence (1) and absence (0) will be shown.
It is possible that the deconvoluted (reconstituted) peak table contains empty features (only 0: absence) following the computational deconvolution step. This might prevent you from using this table in other applications, like NP Analyst, right away. Consult the Cleaning tab for how to remove absent features from the freshly deconvoluted table.
MultiplexMS Demo
A demonstration of the MultiplexMS workflow with detailed instructions can be downloaded here. A copy of the demo instructions can also be found under the demo instructions tab on the left panel.