Welcome
This documentation is intended to clarify the details on how the npmrd_curator is intended to be used, and layout known limitations of the current app.
We recommend keeping note of articles which you encounter which you were not able to complete curation for, and why. This will help us make improvements for v2 of the app.
Please watch the tutorial video on how use the curator app if you have not done so already!
Limitations
1. HTML Tables
There are currently several forms of HTML tables which are likely to fail parsing by our algorithms. In general, the parser is more likely to fail while interpreting 1H NMR data due to extra complexity of these data.
Here are some examples of tables which are known to fail
Tables which contains multiple atom index columns:
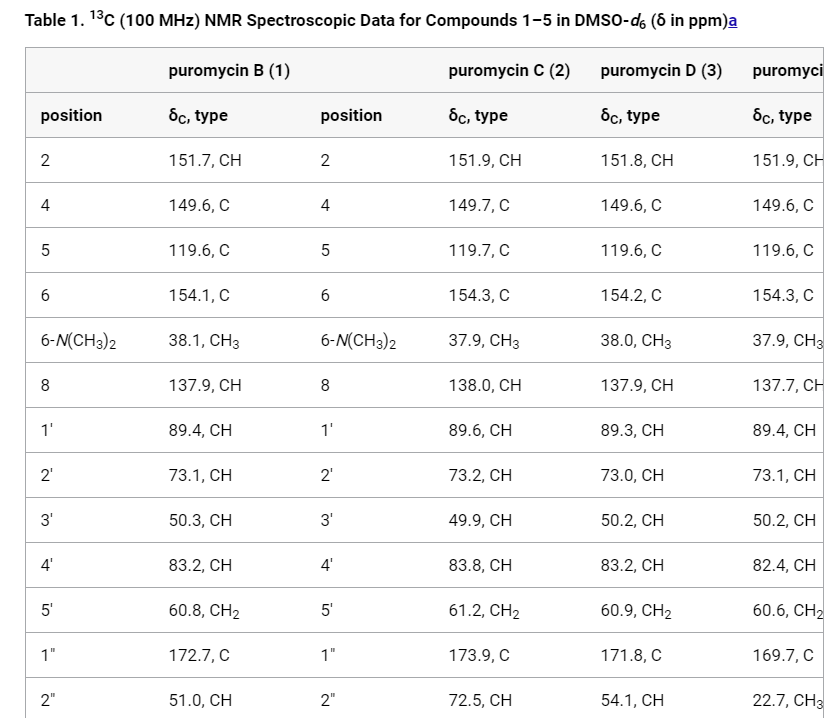
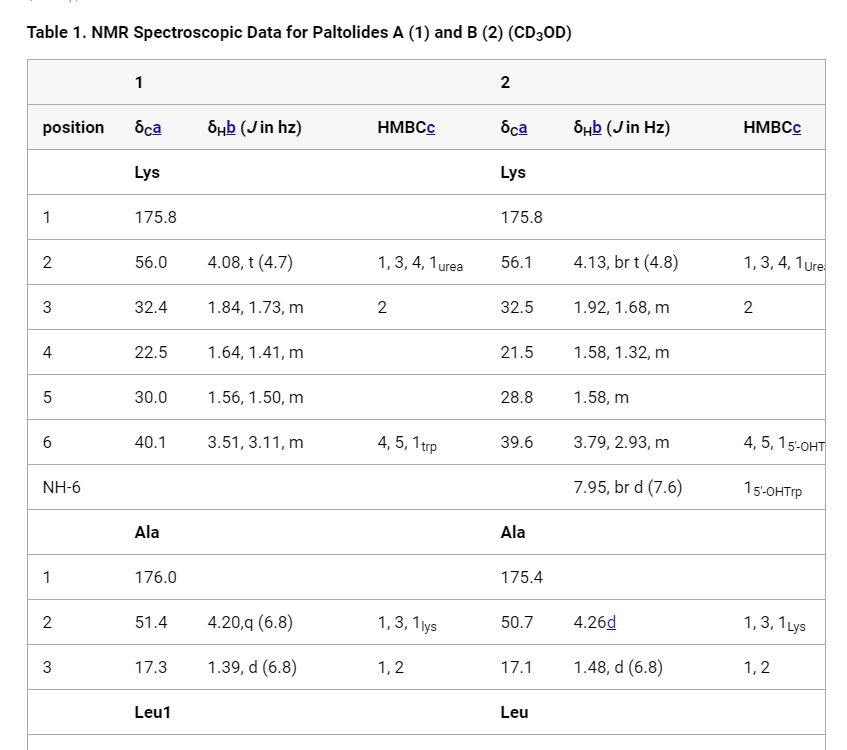
Tables which contain sub-units / residues as a row in the table
Tables which contain integration values may also fail to parse, although these are quite rare.(This limitation has been removed)
Tables which specify two atom indices in a single row will not fail to parse, but you will not be able to properly map literature atom indices to RDKit indices.
1.1 TSV Tables
See Table Parser Page for more details and tips on fixing common problems.
2. Analytical Textblocks
Analytical textblocks are complex and quite difficult to parse 100% accurately. The main culprit for failure during textblock parsing is coupling constants. Please carefully review the “Reconstructed Textblock” to make these data, and all the appropriate metadata were parses correctly.